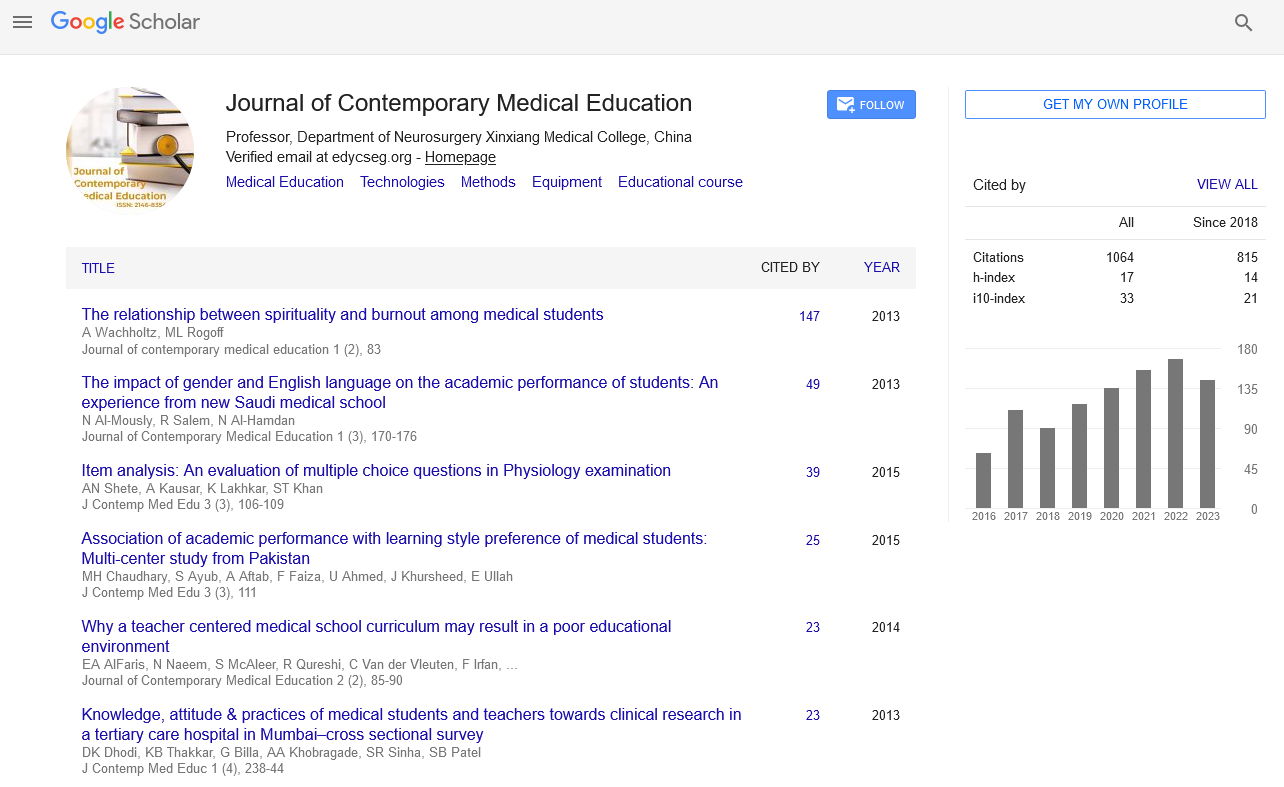
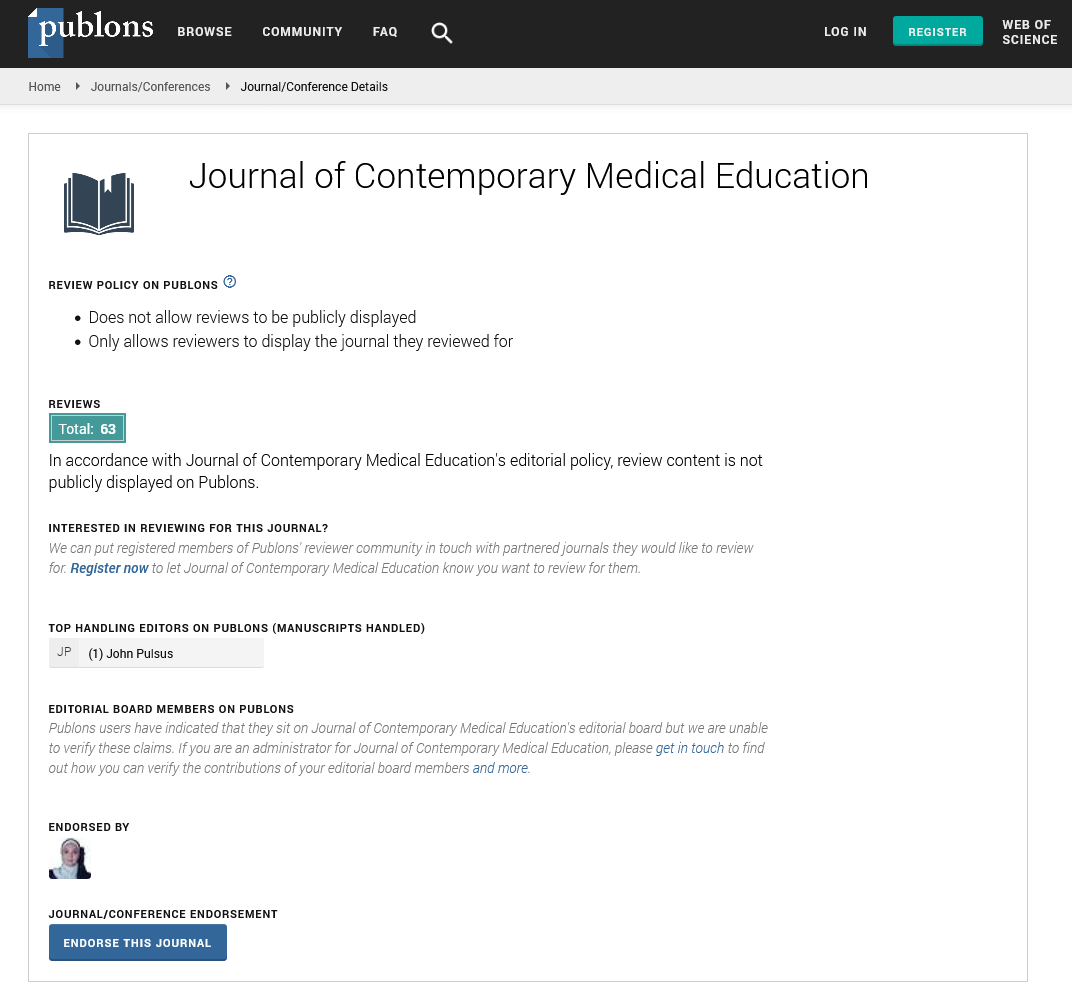
Perspective - Journal of Contemporary Medical Education (2022)
Regulation of Homologous Recombination
Hazm Abas*Hazm Abas, Department of Endodontics, University of the Pacific, California, United States, Email: abaszem@gmail.com
Received: 12-Jul-2022, Manuscript No. JCMEDU-22-72594; Editor assigned: 14-Jul-2022, Pre QC No. JCMEDU-22-72594 (PQ); Reviewed: 28-Jul-2022, QC No. JCMEDU-22-72594; Revised: 04-Aug-2022, Manuscript No. JCMEDU-22-72594 (R); Published: 12-Aug-2022
Description
Homologous recombination is a form of genetic reunification in which the genetic information is exchanged between two identical or nucleic acid molecules with double edges or single strands (usually DNA as in living organisms but may be RNA in viruses). It is widely used by cells to accurately repair the damage that occurs in both DNA strands, known as Double-Strand Breaks (DSB), through a process called Homologous Recombinational Repair (HRR). Homologous regeneration also produces a new set of DNA sequences during meiosis, a process in which eukaryotes produce gamete cells, such as sperm and egg cells in animals. These new DNA sequences represent genetic variation in genes, making it possible for humans to adapt to the evolutionary process. Homologous recombination is also used in horizontal gene transfer to exchange genes between different types and types of bacteria and viruses. Although homologous regeneration varies greatly between living organisms and the different cell types, in double-stranded DNA (dsDNA) most forms involve the same basic steps. After double-stranding of strands, the DNA strands around the 5 ′ ′ end are cut through a process called resection. In the next strand of string attack, à 3 ′ ′ end ends of a broken DNA molecule and then “attacks” the same or similar unbroken DNA molecule. After a string attack, a continuous sequence of events may follow any of the two main mechanisms mentioned below (see Models); DSBR method (Double-Strand Break Correction) or SDSA (route-dependent strand annealing method). Homologous reunification that occurs during DNA repair often leads to incompatible products, actually restoring the damaged DNA molecule as it was before the split of double strands. Homologous reunification has been preserved in all three domains as well as in DNA and RNA viruses, suggesting that it is a virtual biological process. The genetic discovery of homologous reunification in protists by a diverse group of eukaryotic microorganisms has been interpreted as evidence that meiosis arose in early eukaryotic emergence. Since their inactivity is strongly associated with the susceptibility to many types of cancer, proteins make it easier to integrate homologous subjects into active research. Homologous recombination is also used in genetic guidance, a method of introducing genetic mutations into targeted objects. For their development of the program, Mario Capecchi, Martin Evans and Oliver Smithies were awarded the 2007 Nobel Prize for Physiology or Medicine; Capecchi and Smiths independently found applications in embryonic stem cells, however the most conservative methods under the DSB mutation model, which include the same homologous synthetic DNA mutation (gene therapy), were first shown in a plasmid study by -Orr-Weaver, Szostack and Rothstein. Research on plasmid DSB, using γ-irradiation in the 1970-1980s, led to recent experiments using endonuclease (e.g. Homologous Recombination (HR) is important for cell division in eukaryotes such as plants, animals, fungi and protists. In cells differentiated by mitosis, homologous-like regeneration repairs the fragmentation of double strands in DNA caused by ionizing radiation or chemicals that damage DNA. If left untreated, these two-line cuts can cause major chromosomes to regenerate somatic cells, which can lead to cancer. In addition to DNA repair, homologous regeneration also helps to produce genetic diversity when cells divide from meiosis into special gamete cells or sperm cells in animals, pollen or ovules in plants, and seeds in fungi. It does this with the help of a chromosomal crossover, in which the same but different regions of DNA are exchanged between homologous chromosomes. This creates a new, beneficial gene combination, which can give offspring the opportunity to evolve. Chromosomal crossover usually begins when a protein called Spo11 performs the target split of two strands in DNA. These areas are not randomly located on chromosomes; usually in intergenic developer regions and especially in GC-rich domains These areas of duplicate fibers usually occur in regenerative areas, regions with chromosomes that are approximately 1,000–2,000 pairs long and have high regeneration rates. The absence of a recombination hotspot between two genes on the same chromosome usually means that those genes will be passed on to future generations in equal proportions. This represents the interaction between two major genes that would be expected of independent genes during meiosis.
Copyright: © 2022 The Authors. This is an open access article under the terms of the Creative Commons Attribution NonCommercial ShareAlike 4.0 (https://creativecommons.org/licenses/by-nc-sa/4.0/). This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.